-Search query
-Search result
Showing 1 - 50 of 57 items for (author: feldmueller & m)

EMDB-18953: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18954: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M
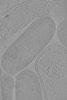
EMDB-18955: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18957: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18958: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18960: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18961: 
Cryo-electron tomogram of mechanically cryo-milled Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-18962: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC cells
Method: electron tomography / : Feldmueller M, Pilhofer M
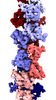
EMDB-18970: 
Subtomogram average of M66 filaments in Yersinia entomophaga cells
Method: subtomogram averaging / : Feldmueller M, Afanasyev P, Pilhofer M
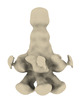
EMDB-18971: 
Subtomogram average of YenTc-Chi2-sfGFP from Yersinia entomophaga chi2-sfGFP
Method: subtomogram averaging / : Feldmueller M, Pilhofer M
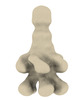
EMDB-18972: 
Subtomogram average of YenTc from Yersinia entomophaga MH96
Method: subtomogram averaging / : Feldmueller M, Pilhofer M

EMDB-19370: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19371: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19372: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19373: 
Cryo-electron tomogram of Yersinia entomophaga delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19374: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19375: 
Cryo-electron tomogram of Yersinia entomophaga chi2-sfGFP cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19376: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta YenTc cells
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19377: 
Cryo-electron tomogram of Yersinia entomophaga MH96 cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M
EMDB-19378: 
Cryo-electron tomogram of Yersinia entomophaga delta LC cells grown at 37 degrees
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19379: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19380: 
Cryo-electron tomogram of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-19381: 
Cryo-electron tomogram of a lysate preparation of Yersinia entomophaga delta LC delta M66 cells
Method: electron tomography / : Feldmueller M, Pilhofer M

EMDB-16284: 
Cryotomogram of L-form-like cytoplasmic membrane vesicles of Listeria monocytogenes Rev2 cell
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16285: 
Cryotomogram of endolysin Ply006 treated Listeria monocytogenes Rev2 cell (stage 1)
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16286: 
Cryotomogram of endolysin Ply006 treated Listeria monocytogenes Rev2 cell (stage 2)
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16287: 
Cryotomogram of a A006 virion attaching to a Listeria monocytogenes Rev2 cell
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16288: 
Cryotomogram of endolysin Ply006 treated Listeria monocytogenes Rev2 cell (stage 3)
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16289: 
Cryotomogram of endolysin Ply007 treated Enterococcus faecalis cells
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16290: 
Cryotomogram of endolysin Ply007 treated Enterococcus faecalis cells
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16291: 
Cryotomogram of individual Efs7 virions and phage attachment to a Enterococcus faecalis cell
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16305: 
Cryotomogram of endolysin Ply007 treated Enterococcus faecalis cells
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-16306: 
Cryotomogram of Listeria monocytogenes Rev2 cells
Method: electron tomography / : Feldmueller M, Wohlfarth JC, Loessner M, Pilhofer M

EMDB-11745: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 3-fold symmetry.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

PDB-7aef: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 3-fold symmetry.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11734: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the cap portion in extended state.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11735: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the sheath-tube module in extended state.
Method: helical / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11743: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 6-fold symmetry.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11744: 
Focused refined cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in extended state applied 6-fold symmetry
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11746: 
Cryo-EM structure of the overall extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11747: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the contracted sheath shell.
Method: helical / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11748: 
Cryo-EM structure of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis, the baseplate complex in contracted state.
Method: single particle / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11749: 
sub-tomogram averaging of an extracellular contractile injection system in marine bacterium Algoriphagus machipongonensis
Method: subtomogram averaging / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-11750: 
sub-tomogram averaging of an extracellular contractile injection system mutant (effector deficient mutant) in marine bacterium Algoriphagus machipongonensis
Method: subtomogram averaging / : Xu J, Ericson C, Feldmueller M, Lien YW, Pilhofer M

EMDB-13704: 
cryo electron tomogram of purified AlgoCIS delta Alg16B
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13705: 
cryo electron tomogram of Algoriphagus machipongonensis in bacterial late stage
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13719: 
Cryo electron tomogram of purified AlgoCIS Cgo1/2 deficient mutant
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13720: 
Cryo electron tomogram of purified AlgoCIS Cgo1 deficient mutant
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13721: 
Cryo electron tomogram of purified AlgoCIS Cgo2 deficient mutant
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M

EMDB-13722: 
Cryo electron tomogram of purified AlgoCIS wild-type.
Method: electron tomography / : Xu J, Ericson C, Feldmueller M, Lien YW, Eisenstein F, Pilhofer M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
